Silvia Onesti
Head of Structural Biology
Higher Education
Master Degree in Chemistry, University of Pavia
PhD in Biophysics (under the supervision of P. Brick & D.M. Blow), Department of Physics, Imperial College London
Previous Appointments
Post-doctoral fellow in Prof. David Blow’s group at Imperial College.
CNR Research Scientist, Department of Genetics, University of Pavia.
Lecturer, Department of Physics, Imperial College.
Senior Lecturer, Department of Biological Sciences, Imperial College.
Contact information
Email: silvia.onesti@elettra.eu, Tel. +39 040 3758451, Mob +39 366 6878001
Elettra - Sincrotrone Trieste S.C.p.A., SS 14 - km 163,5 - AREA Science Park, 34149 Basovizza, Trieste ITALY
Teaching
Teaching at Imperial College:
Molecular Biophysics (Physics); Mechanism of Gene Expression (Biochemistry); Biological Chemistry (Biology); Structural Biology (Master Degree).
Teaching in Trieste:
International School of Advanced Studies (ISAS/SISSA), within the PhD Programs in Physics and Chemistry of Biological systems, Structural and Functional Genomics and Neurobiology; Master degree in Functional Genomics (University of Trieste).
Various
Member of the Editorial Board of Scientific Reports, a new open access publication from Nature Publishing Group, covering all areas of natural sciences (http://www.nature.com/srep/about/index.html).
Chairman of the of INSTRUCT Italian Working Group on Complementary Techniques
(http://www.cerm.unifi.it/about-cerm/italian-users-of-instruct).
Member of the IUCr Commission on Biological Macromolecules 2009-2015 (http://www.iucr.org/iucr/commissions/cbm.html).
Member of the International Programme Committee for the organization of the IUCR 23rd Congress and General Assembly to be held in Montreal 5-12 August 2014. (http://iucr2014.org/side_organization/international_program_committee_e.shtml).
Beneficiary of the AntiHelix MSCA-ETN (https://www.elettra.eu/AntiHelix/Elettra/HomePage).
Member of the CAPSTONE MSCA-ETN Scientific Advisory Committee (http://www.capstone-etn.eu/).
Structural biology is an interdisciplinary research area, requiring expertises from both the life sciences and the physical sciences. We apply molecular and structural biology tools to study the basic genetic processes within the cell, such as DNA replication and transcription. We use protein crystallography to determine the atomic structure of eukaryotic and archaeal proteins involved in these processes. Crystallographic studies are complemented by the concomitant use of electron microscopy to visualise the architecture of large complexes.
DNA replication, transcription and DNA repair are crucial event in the cell cycle, underpinning cellular processes with important consequences such as cell proliferation and genome stability. Failure to control these processes causes chromosome instability, which can lead to the development of cellular abnormalities, genetic disease and the onset of cancer.
Structural studies of aminoacyl-tRNA synthetases
Aminoacyl-tRNA synthetases play a crucial role in the fidelity of translation of the genetic code by catalyzing the attachment of an amino acid to the specific tRNA molecule. I have obtained the first detailed information on the structure of lysyl-tRNA synthetase, in a complex with the substrate lysine (Onesti et al., 1994; Onesti et al., 1995). I have also determined the structures of a series of different enzyme-ligand complexes which provide detailed information on the induced conformational changes of the enzyme as well as on the interactions made with ATP and the aminoacyl-adenylate intermediate (Desogus et al, 2000; Onesti et al., 2000).
Structural studies of RNA polymerase subunits & other proteins involved in transcription
RNA polymerases are at the core of the complex network that controls gene expression, and thus the fate of each cell. We have determined the crystal structure of the evolutionary conserved subunit RPB5 (Todone et al., 2000) and the archaeal and human RPB4/RPB7 heterodimer that reversibly associates with the RNA polymerase II core (Todone et al., 2001; Meka et al., 2003)). When the crystal structure of the entire RNA polymerase was determined, our structures were fitted into the low-resolution electron density, to help model building. We have also detected a structural and functional homology between RPB4/RPB7 and the RNA polymerase I subunits A14/A43 (Meka et al., 2005). We have also characterized the Elp3 subunit of the transcription Elongator complex, and particularly the Radical SAM domain, showing that it contains a Fe4S4 cluster and binds S‑adenosyl-methionine (Paraskevopoulou et al., 2006). We have expressed and purified various subcomplexes of the yeast and human Mediator complex.
Structural studies of MCM proteins
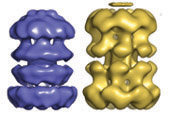
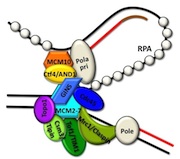
A key component of the replisome is the MCM replicative helicase, opening up the DNA double helix ahead of the replication fork. We have used a combination of electron microscopy and crystallography to understand the architecture of a simplified archaeal MCM complex and have assessed the changes in stoichiometry in presence of various substrates and/or mutations (Pape et al., 2003; Pucci et al, 2004; Costa et al., 2006a; Costa et al., 2006b; Bae et al. 2009; Jenskinson et al., 2009). We have also visualized the initial interaction between MCM and dsDNA, prior to the loading and activation of the complex to function as a helicase at the fork (Costa et al. 2008). Due to these key results, I have been invited to write various reviews and book chapters, on MCM helicases (Costa & Onesti, 2009; Costa & Onesti, 2009) and on hexameric helicases (Medagli & Onesti 2013).
Structural studies of other DNA replication proteins
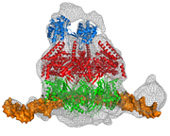
Cdc45, MCM and GINS form the CMG complex, which acts as a replicative helicase. We have used a combination of electron microscopy and small-angle X-ray scattering to resolve the controversy regarding the structure of the GINS complex (Carroni et al. 2017). Cdc45 is an essential protein conserved in all eukaryotes and is involved in both the initiation of DNA replication and the progression of the replication fork. We detected a weak but significant relationship among eukaryotic Cdc45 proteins and a large family of phosphoesterases that has been described as DHH family (Krastanova et al., 2012). More recently, we have determined the crystal structures of two archaeal homologues of Cdc45, with and without nucleic acid substrates, and characterized them biochemically (manuscripts in preparation). Based on our results, I have been invited to write a review (Onesti and MacNeill, 2013) and a book chapter (Medagli et al., 2016) on the CMG complex.
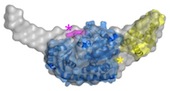
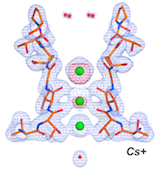
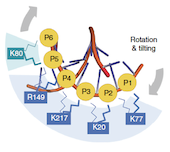
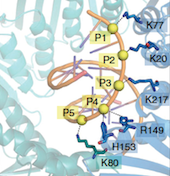
We have cloned, expressed and biochemically characterized the primase from Methanococcus jannaschii, the first primase from archaeal organisms (Desogus et al. 1999). We have also carried out structural studies of PCNA in complex with DNA (De March et al., 2017) and DNA/p15PAF (De March et al., 2018), obtaining a structural picture of the clamp sliding mechanism and the role of p15PAF in DNA replication, as well as a picture of how non-canonical PIP motifs bind to the ring (Gonzalez-Magaña et al., 2019).
Functional and structural studies on RecQ and FeS helicases
RecQ4 helicase is an important member of the human RecQ family that plays a role in DNA replication and DNA repair. It is involved in three rare genetic diseases as well as in sporadic and genetic cancers. We carried out a bioinformatics analysis that revealed the presence of novel domains (Marino et al. 2013). We determined the NMR structure of a Zn knuckle located at the N-terminus and showed that it binds to both DNA and RNA substrates (Marino et al., 2016), and biochemically characterized the catalytic core (Mojumdar et al. 2016). We have recently measured the activity towards a variety of unusual nucleic acid structures (manuscript in preparation). We have recently started to work on a number of helicases containing a FeS cluster, including DinG, DDX11 and RTEL1 (Pisani et al., 2018; Bottega et al., 2019). In collaboration with the NanoInnovation Laboratory, we developed a new cheap and robust diagnostic method to measure helicase activity, that can be easily implemented for high-throughput screening of potential helicase inhibitors (Deka et al., 2017).
Outreach
- 25/11/2022: Parola di Chimica! Orientarsi tra le scienze della vita. Webinar AIRC per le scuole superiori, con Riccardo Rollini.https://webinarscuola.airc.it/webinar/81 https://www.youtube.com/watch?v=c02u6MZ4hZk
- 18/10/2022: Intervistata dalla giornalista scientifica Simona Regina per RADAR, trasmissione radiofonica, per commentare i premi Nobel 2022 https://t.co/0bgtj1vECg
- 18/05/2021: "Vedere l'invisibile: come come la biologia strutturale aiuta la ricerca oncologica" AIRC Campus, Università dell'Insubria, ospite di Viviana Orlandi, Rosalba Gornati e Giorgio Binelli. https://campus.airc.it/progetto/varese-universita/
- 03/01/2021: Treccani Atlante, Il Faro: La fiducia nella scienza durante la pandemia https://www.treccani.it/magazine/atlante/societa/La_fiducia_nella_scienza_durante_la_pandemia.html
- 26/11/2020: "Tra verità e incertezza: la cultura scientifica in Italia" in "Scienza e scelte politiche durante il Covid-19", AAVV https://www.amazon.it/dp/B08NSVVTZP/ref=sr_1_1?
- 25/09/2020: Trieste Next 2020 - Il tallone d’Achille: bersagli molecolari per terapie antivirali, antibatteriche e anticancro. https://www.triestenext.it/archives/gw_relatori/silvia-onesti
- 03/09/2020. Science and the City Festival/ESOF2020 - #hofiducianellaricerca: i ricercatori AIRC dai loro laboratori di Trieste raccontano come garantire continuità alla ricerca oncologica. Silvia Onesti e Gianni del Sal al Caffè San Marco. https://events.scienceinthecity2020.eu/it/hofiducianellaricerca-i-ricercatori-airc-dai-loro-laboratori-di-trieste-raccontano-come-garantire-continuita-alla-ricerca-oncologica
- 02/09/2020: RaiRadioTre - puntata dedicata a Elettra e Area Science Park. Con Giorgio Paolucci, Silvia Onesti, Andrea Lausi, Steve Taylor, https://bit.ly/3nd2b3b
- 13/06/2020: Tales of World Scientists https://www.facebook.com/watch/?v=682657185627944
- 10/06/2020: Treccani Atlante: Parole oltre la pandemia: Scienza https://www.treccani.it/magazine/atlante/societa/Scienza_parole_pandemia.html
- 22/05/2020: Festival Digitale Treccani - A round table on "Science and information during the pandemic" - with Gaetano Savatteri, Silvia Onesti, Claudio Cartoni, Andrea Carlino. https://www.facebook.com/watch/?v=3128572413832237
- 15/05/2020: AIRC Campus - lecture to Pharmaceutical Chemistry students, University of Naples. https://campus.airc.it/airc/ http://www.farmacia.unina.it/-/21998961-lezione-airc-sivia-onesti;jsessionid=B856B3614EAB3825325579E0B8217795.unina_dip2
- 13/05/2020: Risorgimento Digitale - Maestri D'Italia: Vedere l'invisibile https://timgate.it/video/risorgimentodigitale/silvia-onesti-vedere-l-invisibile.vum
- 19/04/2020: Treccani Atlante, Il Faro: La rivincita dei saperi https://www.treccani.it/magazine/faro/edizioni/il-faro-19-4-20.html
- 02/04/2020: Lockdown in Italy: personal stories of doing science during the COVID-19 quarantine. Nature - Career features. https://www.nature.com/articles/d41586-020-01001-8
- 07/09/2019: Giornata mondiale della ricerca sul cancro - TGR visit to the laboratory. https://www.rainews.it/dl/rainews/TGR/multimedia/ContentItem-2a894f50-7a0f-40e6-b6c7-1cf358fd73e8.html
- 27/09/2019: Trieste Next 2019 - Sistemi periodici, dialogo tra Chimica e Letteratura. https://disu.units.it/it/eventi/37318
- 28/09/2019: Trieste Next 2019 - Il futuro della ricerca medica tra app e nuove tecnologie. Tavola rotonda con Massimo Temporelli, Cristiano Petrini e Silvia Onesti https://www.facebook.com/watch/live/?v=879518519096053
- 14/03/2017: Lectio magistralis Treccani: Fare luce sulle proteine. https://www.treccani.it/magazine/webtv/videos/Conv_Onesti_Silvia.html
- 25-27/09/2015 Trieste Next 2015: Stand "Facciamo luce sulle Proteine" https://www.areasciencepark.it/events/le-eccellenze-del-sistema-area-a-trieste-next-dal-25-al-27-settembre/ https://www.elettra.trieste.it/comunicazione/events/elettra-a-trieste-next-2015.html
- 26/09/2015: Trieste Next 2015 - Srotolare il DNA https://www.elettra.trieste.it/comunicazione/events/elettra-a-trieste-next-2015.html
- 07/05/2011: NITLAB in AVVENTURA DELLA SCIENZA, Milano. Caffe’ scientifico con rinfresco e musica: Le donne nella Scienza con Silvia Onesti (Sincrotrone, Trieste) e Caterina La Porta (UniMi) http://www.nitlab.unimi.it/avventure2011.html
- 10/01/2010: Sincrotrone, tre sfide anticancro https://contao.icgeb.org/tl_files/Notizie_2012/Notizie2012pdf/MICROSCOPIO%202012/Microscopio1.pdf
Funding
Associazione Italiana per la Ricerca sul Cancro (AIRC: http://www.airc.it/)
- IG 20778: Understanding the role of RecQ4 in cancer development and progression
- IG 14718: The human CMG helicase in 3D: structural and functional studies on the single components and the assembly of the complex
- IG 10646: "Elucidation of the structure and function of human MCM helicases
Programma INTERREG Crossborder Cooperation Programme Italy-Slovenia 2007-2013 & 2014-2020
French Muscular Dystrophy Association (AFM Téléthon)
H2020 Marie Skłodowska Curie Action ITN (MSCA-ITN http://www.elettra.eu/AntiHelix/)
Recent publications
(*Corresponding author)
Atomic Force Microscopy investigation of the interactions between the MCM helicase and DNA Mohammed Khalid A.A., Parisse P.*, Medagli B., Onesti S.*, Casalis L.* (2021). Materials 14, 687.
Mitochondrial defect in Warsaw syndrome cells genomic integrity and mitochondrial metabolism defects in Warsaw syndrome cells: A comparison with Fanconi anemia.
Bottega R., Ravera S., Napolitano L.M.R., Chiappetta V., Zini N., Crescenzi B., Arniani S., Faleschini M., Cortone G., Faletra F., Medagli B., Sirchia F., Moretti M., de Lange J., Cappelli E., Mecucci C., Onesti S., Pisani F.M., Savoia A.* (2021). J Cell Physiol. Online ahead of print.
Reply to: "Does PCNA diffusion on DNA follow a rotation-coupled translation mechanism?
De March M., Onesti S*. and De Biasio A.* (2020). Nat. Commun. 11, 4999.
C-Terminal Domain of the Human Zinc Transporter hZnT8 Is Structurally Indistinguishable From Its Disease Risk Variant (R325W).
Ullah R., Shehzad A., Ali Shah M., De March M., Ismat F., Iqbal M., Onesti S., Rahman M., McPherson M.J.* (2020). Int J Mol Sci, 21, 926.
Two further patients with Warsaw breakage syndrome. Is a mild phenotype possible?
Bottega R., Napolitano L.M.R., Carbone A., Cappelli E., Corsolini F., Onesti S., Savoia A., Gasparini P., Faletra F.* (2019). Mol. Genet. Genom. Med. 7, e693.
Analysis of the Zn-binding domains of TRIM32, the E3 ubiquitin ligase mutated in Limb Girdle Muscular Dystrophy 2H.
Lazzari E., El-Halawany M., De March M., Valentino F., Cantatore F., Migliore C., Onesti S., Meroni G.* (2019). Cells, 8, E254.
The p12 subunit of human polymerase δ uses an atypical PIP-box for molecular recognition of proliferating cell nuclear antigen (PCNA).
Gonzalez-Magaña A., Ibáñez de Opakua A., Romano-Moreno M., Murciano-Calles J., Merino N., Luque I., Rojas A.L., Onesti S., Blanco F.J., De Biasio A.* (2018). J. Biol. Chem. 294, 3947-3956.
Molecular and Cellular Functions of the Warsaw Breakage Syndrome DNA Helicase DDX11.
Pisani F.M.*, Napolitano E., Napolitano L.M.R. and Onesti S.* (2018). Genes 9, 564.
p15PAF binding to PCNA modulates the DNA sliding surface.
De March M., Barrera-Vilarmau S., Crespan E., Mentegari E., Merino N., Gonzalez-Magaña A., Romano-Moreno M., Maga G., Crehuet R., Onesti S., Blanco F.J. and De Biasio A.* (2018). Nucleic Acids Res. 46, 9816-9828.
The permeation mechanism of organic cations through a CNG mimic channel.
Napolitano L.M.R., Marchesi A., Rodriguez A., De March M., Onesti S., Laio A.* and Torre V..* (2018). PLoS Comput. Biol. 14, e1006295.
DNA-conjugated gold nanoparticles based colorimetric assay to assess helicase activity: a novel route to screen potential helicase inhibitors.
Deka J., Mojumdar A., Parisse P., Onesti S.* and Casalis L.* (2017). Scientific Rep. 7, 44358
Structural basis of human PCNA sliding on DNA.
De March M., Merino N., Barrera-Vilarmau S., Crehuet R., Onesti S*., Blanco F.S*. and De Biasio A.* (2017). Nat. Commun. 7, 13935.
New insights into the GINS complex explain the controversy between existing structural models.
Carroni M., De March M., Medagli B., Krastanova I., Taylor I.A., Amenitsch H., Araki H., Pisani F.M., Patwardhan A. and Onesti S.* (2017). Scientific Rep. 7, 40188.
The human RecQ4 helicase contains a functional RQC domain that is essential for activity.
Mojumdar A., De March M., Marino F. and Onesti S.* (2016). J. Biol. Chem. 292, 4176-4184.
The tumor suppressor ING4 binds double stranded DNA with micromolar affinity through its disordered central region.
Ormaza G., Medagli B., Rodríguez J.A., Ibáñez de Opakua A., Merino N., Villate M., Onesti S. and Blanco F.J.* (2016). FEBS Letters. 591, 425-432.
Structure and activity of the Cdc45-Mcm2-7-GINS (CMG) complex, the replication helicase.
Medagli B., Di Crescenzio P., De March M. and Onesti S.* (2016). (Chapter in The initiation of DNA replication in eukaryotes, Ed. D. Kaplan, Springer).
Structural and biochemical characterization of an RNA/DNA binding motif in the N-terminal domain of RecQ4 helicases.
Marino F., Mojumdar A., Zucchelli C. Bhardwaj A., Buratti E., Vindigni A., Musco G. and Onesti S.* (2017). Scientific Rep. 6, 21501.
Status of the crystallography beamlines at Elettra.
Lausi A.*, Polentarutti M., Onesti S., Plaisier J.R., Busetto E., Bais G., Barba L., Cassetta A., Campi G., Lamba D., Pifferi A., Mande S.C., Sarma D.D., Sharma S.M., Paolucci G. (2015). Eur. Phys. J. Plus 130, 43-51.
A structural, functional, and computational analysis suggests pore flexibility as the base for the poor selectivity of CNG channels.
Napolitano L.M.R., Bisha I., De March M., Marchesi A., Arcangeletti M., Demitri N., Mazzolini M., Rodriguez A., Magistrato A., Onesti S.*, Laio A.* and Torre V.* (2015). Proc. Natl. Acad. Sci. USA 112, E3619-E3628.
Bioinformatic analysis of RecQ4 helicases reveals the presence of a RQC domain and a Zn knuckle.
Marino F., Vindigni A. and Onesti S.* (2013). Biophys Chem. 177-178, 34-39.
Structure and evolutionary origins of the CMG complex.
Onesti S. and MacNeill S.A.* (2013). Chromosoma 122, 47-53.
Structural and functional insights into the DNA replication factor Cdc45 reveal an evolutionary relationship to the DHH family of phosphoesterases.
Krastanova I., Sannino V., Amenitsch H., Gileadi O., Pisani F.M., Onesti S.* (2012). J. Biol. Chem. 287, 4121-4128.
Structural biology of MCM helicases.
Costa A. and Onesti S.* (2009). Crit. Rev. Biochem. Mol. Biol. 44, 326-342.
Cryo-electron microscopy reveals a novel DNA binding site on the MCM helicase.
Costa A., Van Dujinen G., Medagli B., Chong J., Sakakibara N., Kelman Z., Nair S.K., Patwardhan A. and Onesti S.* (2008). EMBO J. 27, 2250-2258.
Structural basis of the Methanobacter thermautotrophicus MCM helicase activity.
Costa A., Pape T., van Heel M., Brick P., Patwardhan A. and Onesti S.* (2006)
Nucleic Acid Res. 34, 5829-5838.
The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine.
Paraskevopoulou C., Fairhurst S.A., Lowe D.J., Brick P. and Onesti S.* (2006)
Mol. Microbiol. 59, 795-806.
Crystal structure and RNA binding of the Rpb4/Rpb7 subunits of human RNA polymerase II.
Meka H., Werner F., Cordell, S., Onesti S. and Brick P.* (2005)
Nucleic Acid Res. 33, 6435-6444.
Hexameric ring structure of the full-length archaeal MCM complex.
Pape T., Meka H., Chen S., Vicentini G., van Heel M. and Onesti S.* (2003)
EMBO Rep. 4, 1079-1083.
Structural and functional homology between the RNAPI subunits A14/A43 and the archaeal RNAP subunits E/F.
Meka H., Daoust G., Bourke-Arnvig K., Werner F., Brick P. and Onesti S.* (2003)
Nucleic Acid Res. 31, 4391-4400.
Structure of an archaeal homologue of the eukaryotic RNA polymerase II RPB4/RPB7 complex.
Todone F., Brick P., Werner, F., Weinzierl R.O.J and Onesti S.* (2001)
Mol. Cell, 8, 1137-1143.
Crystal structure of RPB5, a universal eukaryotic RNA polymerase subunit and transcription factor interaction target.
Todone F., Weinzierl R.O.J, Brick P. and Onesti S.* (2000)
Proc. Natl. Acad. Sci. USA, 97, 6306-6310.